Vanishing beasts: the conservation of African rhinoceros
Event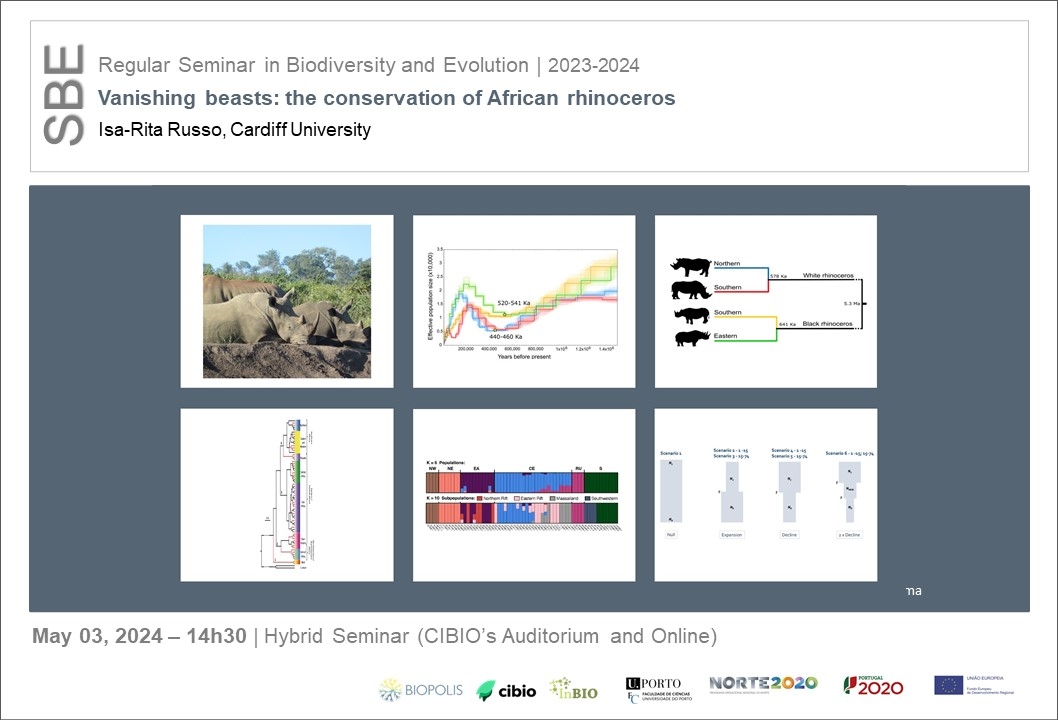
REGULAR SEMINAR IN BIODIVERSITY AND EVOLUTION
May 03rd, 2024
Isa-Rita Russo, Cardiff University | 14h30 | Hybrid Seminar

REGULAR SEMINAR IN BIODIVERSITY AND EVOLUTION
Molecular tools are used for the assessment and monitoring of biodiversity which aid in conservation decisions and management. Recent developments in molecular methods have allowed researchers to analyse species at a genetic and/or genomic level. The level of analysis will depend on the available resources, funding, training, and access to technologies. In some countries, traditional genetic data are still utilised in conservation decisions and in others advanced technologies that produce genome data for species are incorporated into management. Genetic/genomic information can answer a variety of research questions at either a higher evolutionary level or at phylogeographic, phylogenetic or population level. Here I have used African rhinoceros as a case study to illustrate how genetic and genomic information can complement each other in conservation management. Genetic (microsatellites and mitochondrial DNA) data for black rhinoceros showed erosion of genetic diversity across the distributional range of the species and several evolutionarily significant units (ESUs) have been identified for the species. For white rhinoceros, two genetically distinct lineages/clusters were identified which correspond to the two recognised white rhinoceros subspecies. In addition, both subspecies have undergone recent population size changes. Results also indicated interspecific gene flow between the precursor species of both white and black rhinoceros until about 3.3 to 4.1 MYA. Data also showed that genome level analysis often identified finer scale patterns that were not observed using traditional markers (mitochondrial DNA or microsatellite data), but the overall patterns of diversity were supported by all marker types. In addition, genomic data provided further information regarding inbreeding and genetic load. Researcher should therefore choose suitable molecular makers according to the research questions, conservation goals for the species and the available funding.
I am a population geneticist with experience in population and landscape genetics/genomics. My research focusses on how landscape variables affect gene flow and adaptation in several taxa (brown bears, giant pandas and small mammals, African buffalo) by combining large-scale genetic, disease and environmental data sets to test for correlations between landscape features/disease status and gene flow/adaptation. I am also interested in how genetic variation has been shaped due to past demographic events and changes in population size using Approximate Bayesian Computation and pairwise sequentially Markovian coalescent methods for microsatellite/SNP and whole genome sequencing data respectively. In addition, I am involved in several projects as a member of the IUCN Conservation Genetics Specialist Group (CGSG) at the policy-science interphase. As one of the co-coordinators of the African node of CGSG I am leading a project aiming to collect, collate and interpret the current knowledge of the genetic composition of southern African vertebrates and make this information accessible to conservation practitioners, the wildlife industry, and governments as genetic management guidelines.
[Host: Maria Joana Silva, Biodiversity of Deserts and Arid Regions - BIODESERTS]
